Research Highlights as Told by Our Early Career Scientists
Research Highlights as Told by Our Early Career Scientists
We asked our students and post-docs to tell us about their latest publications...
UnidecNMR: Automatic Peak Detection for NMR Spectra in 1-4 Dimensions
Here at the Kavli Institute for Nanoscience Discovery, we use a variety of techniques to visualise the world at nanoscopic scale, including Mass Spectrometry and Nuclear Magnetic Resonance (NMR). A core aim for our institute is to create bridges between researchers who use these different methods, so that we can tackle the most complex nanoscience problems from multiple perspectives. However, each technique produces different, complex data, requiring careful interpretation. Many of our research groups try to simplify this by building easy-to-use data analysis tools.
A few years ago, one of our Mass Spectrometry focused groups (the Robinson Lab), carried out research that resulted in the Unidec algorithm, which has since proven instrumental in understanding the behaviour of proteins. Now, a neighbouring group (the Baldwin Lab) has adapted this approach for a different method, resulting in UnidecNMR, a recently published tool which simplifies complex NMR data analysis. UnidecNMR has already helped Baldwin group researchers unveil how the SARS-CoV-2 Spike protein interacts with the sugars that coat our cells, and the authors are hopeful that their tool will enable deeper nanoscopic research worldwide.
Professor Andrew Baldwin commented: “The Unidec algorithm is a great tool that enables researchers to use the leading methods such as EM, NMR and MS to better understand biology.”
The publication can be read here
Written by Dr Charles Buchanan, Alumni from the Department of Chemistry and Kavli Oxford.
Charles completed their DPhil with Professor Andrew Baldwin
Single extracellular vesicle detection assay identifies membrane-associated α-synuclein as an early-stage biomarker in Parkinson's disease
Extracellular vesicles (EVs) are tiny particles released by cells that carry distinctive proteins reflecting their origin, including those linked to disease. They are increasingly recognised as promising biomarkers for conditions such as Parkinson’s disease (PD). However, most current EV-based tests rely on bulk protein measurements, which can be affected by contaminants and overlook signals from rare, but disease-relevant EV subtypes. In this study, we developed a microfluidic platform that enables parallel analysis of individual EVs that are compartmentalised in oil droplets, in this way allowing precise quantification of surface proteins in specific EV subpopulations. Using this technology, they focused on L1CAM-positive EVs (L1EVs), a rare EV subtype derived from neurons and released in the circulation. We discovered that α-synuclein, a key protein implicated in PD, is enriched on the surface of L1EVs and that this form is partly pathological, as evident by detection with antibodies that recognise aggregated forms of this protein and confirmed using disease-relevant cellular models including patient stem cell-derived neurons with PD-associated mutations. Importantly, membrane-associated α-synuclein was detected with high diagnostic accuracy in individuals who are at risk of developing PD (AUC=0.93), and in patients with clinically diagnosed PD (AUC=0.95). These findings suggest that L1EV surface α-synuclein in blood is a highly promising, non-invasive biomarker for early and accurate PD diagnosis. More broadly, this single-EV resolution approach could transform biomarker discovery across neurological diseases by enabling detailed profiling of rare EVs directly from patient samples.
The publication can be read here.
Written by Shijun (Victor) Yan, Doctoral student, Nuffield Department of Clinical Neurosciences and Kavli Oxford with Professor George Tofaris
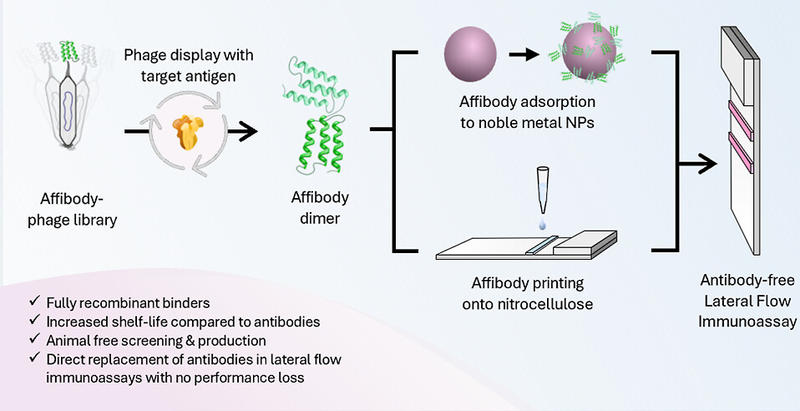
Adding a Twist to Lateral Flow Immunoassays: A Direct Replacement of Antibodies with Helical Affibodies, from Selection to Application
Lateral flow immunoassays (LFIAs) are a class of rapid diagnostic test used to diagnose a range of diseases, and were implemented widely during the Covid-19 pandemic. LFIAs typically use antibodies to bind to disease-specific targets. However, the use of antibodies comes with a number of limitations, including batch-to-batch variability, use of animal models in production, and limited stability.
In this work, traditional antibodies were replaced with engineered affinity proteins, known as affibodies. Affibodies are a class of small, helical proteins that can be selected to bind to disease-specific target proteins. Here, affibodies were produced to bind to the SARS-CoV-2 spike protein, and screened to generate a library of suitable affinity agent pairs.
Affibodies were incorporated into LFIAs as a like-for-like replacement to antibodies, following industry-standard manufacturing methods for surface functionalisation, to ensure that the incorporation of affibodies did not pose a significant barrier to large-scale LFIA manufacturing. The affibody-based LFIAs demonstrated comparable performance to traditional antibody-based LFIAs, and were compatible for use with human samples, such as saliva.
The incorporation of affibodies into LFIAs offers several key advantages. Specifically, due to their sequence-definition, affibodies were readily modified to produce multimeric affinity agents, increasing binding affinity. Further, the enhanced stability of affibody constructs was demonstrated, where affibody-based LFIAs showed improved stability at elevated humidity and temperature when compared to antibody analogues. This finding is particularly beneficial for the development of diagnostic tests for use in resource-limited setting.
The publication can be read here
Written by Christy Sadler, Doctoral student, Department of Physiology, Anatomy and Genetics and Kavli Oxford with Professor Dame Molly Stevens
Ribosome phenotypes for rapid classification of antibiotic-susceptible and resistant strains of Escherichia coli
Ultra-rapid antibiotic susceptibility tests (ASTs) that can return results within a few hours are increasingly important for treating antibiotic-resistant infections and for minimising antibiotic use. Antibiotic susceptibility phenotyping on a single-cell basis, through the visualisation of intracellular changes associated with the antibiotic response, is a promising technology for ASTs because it could eliminate lengthy bacterial culturing steps and provide a rich image dataset to study resistance mechanisms and bacterial heterogeneity. These data can be used to train neural networks to identify phenotypes associated with antibiotic resistance or susceptibility. In this paper, we extend these methods by showing that the spatial distribution of fluorescently labelled ribosomes, or the ribosome phenotype, can be used as an AST. We show that ribosomes are a useful target for single-cell phenotyping because they are abundant, outline the cell periphery for image segmentation, and act as an inverse DNA stain. Ribosomes also contain RNA sequences that can be used for species identification, if species-specific probes are used. We show that a neural network can use the ribosome phenotype to identify an antibiotic response for four clinically relevant classes of antibiotics, with different mechanisms of action, with accuracies >95%. We then apply the method to E. coli strains isolated from patients with bloodstream infections and show that it can classify ciprofloxacin-susceptible cells with 91% accuracy and resistant cells with 99% accuracy. With these accuracies, we predict that our method could differentiate a resistant sample from a susceptible one with 99% confidence with as few as 2 cells.
The publication can be read here
Written by Dr Alison Farrar, Alumni from the Department of Physics and Kavli Oxford.
Alison completed their DPhil with Professor Achillefs Kapanidis


